Genome Browser Integration, Related Datasets and More New Features on FaceBase
Published 28 January 2020
We have updated the Data Browser by integrating the UCSC Genome Browser directly in the record pages, including a section of related datasets (providing the ability to link to related FaceBase datasets and external sources such as dbGap or GEO) and upgrading the look and feel.
UCSC Genome Browser Integration
For sequencing datasets that include track data, FaceBase updates our trackhub nightly with any newly released track data. We also integrate the GB visualization right in the dataset’s record page.
For example, the screenshot below shows the detail page for the ChIP-seq of multiple histone marks and RNA-seq from e11.5 mouse face subregions dataset. You can see the Genome Browser section below the Summary.
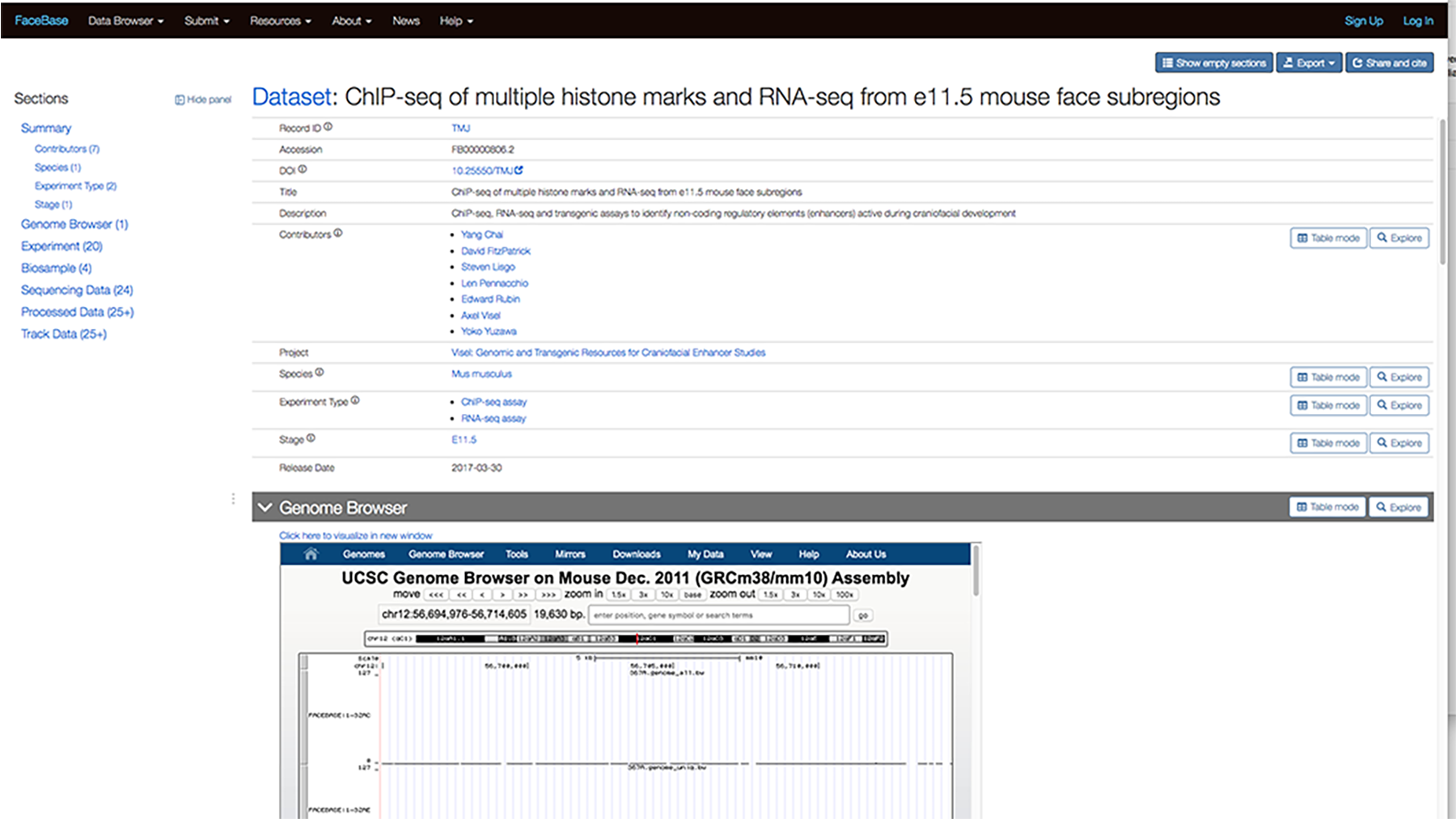
You can scroll down further to view the data while still on the page.
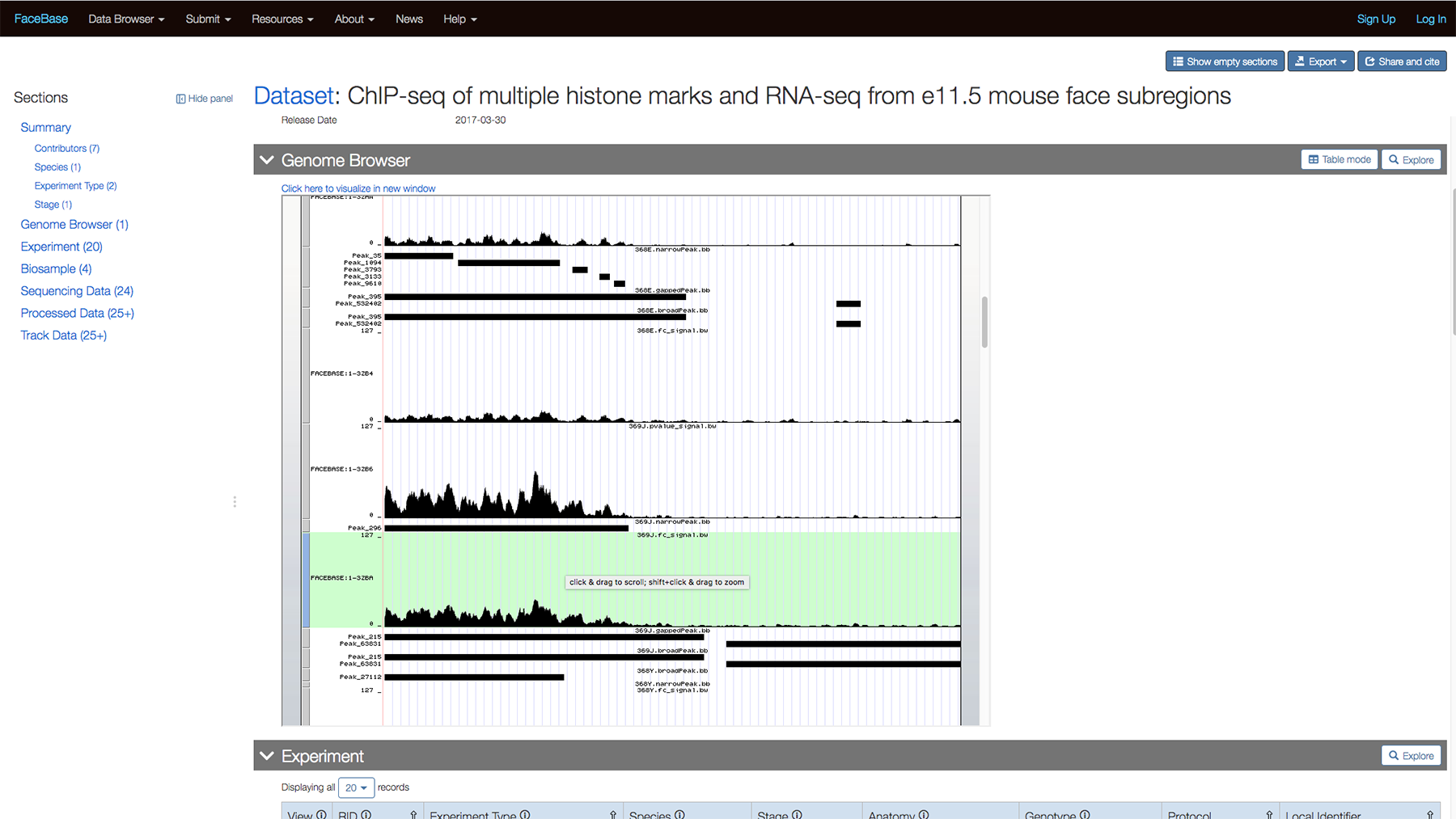
Or click “Click here to view visualization in new window” to view the track on the UCSC Genome Browser.
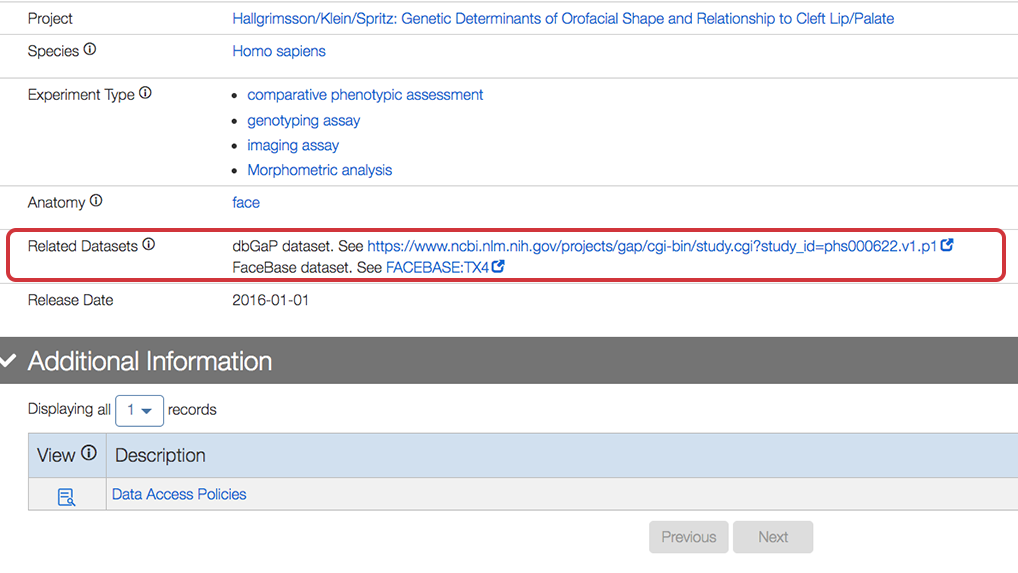
Related Datasets section
We can now indicated datasets that are related to each other, whether in FaceBase or another repository such as dbGAP via the “Related Datasets” section. The following example is from the detail page for Sample to subject mapping file for the 3D Facial Images-Tanzania dataset.
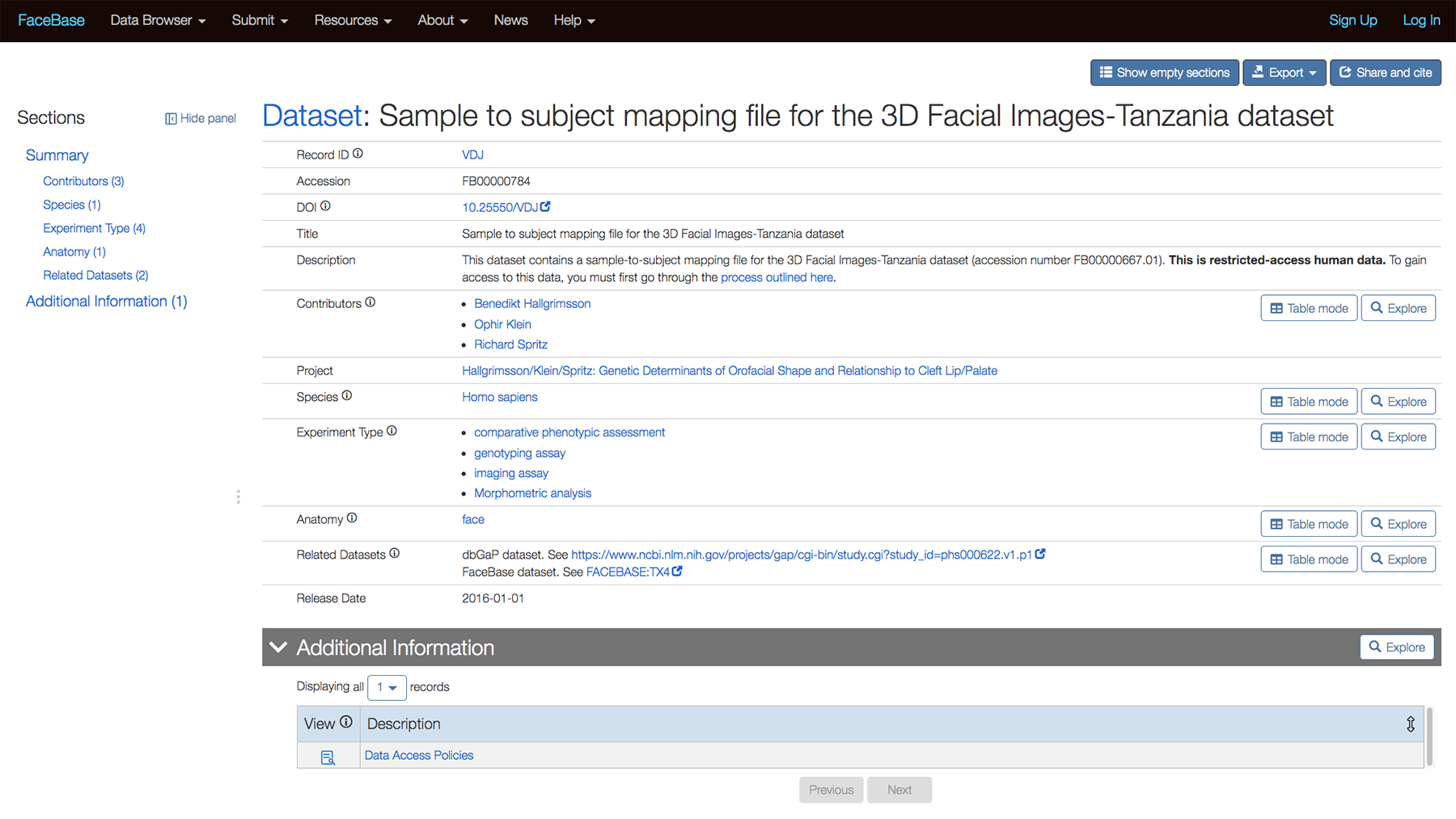
The following is a closeup of the entries for the Related Datasets section:
Updated User Interface for the Data Browser
As a result of extensive usability testing, we’ve made some foundational changes to the style and layout of the Data Browser as you can see in the above screenshots. These changes focused on making content in the Data Browser easier to read and to standardize layout and formatting elements. For example, the section headers on a detail page are much more prominent, more distinguishable from the rest of the content and easier to scan.
