Announcing the release of MusMorph
Published 09 November 2021
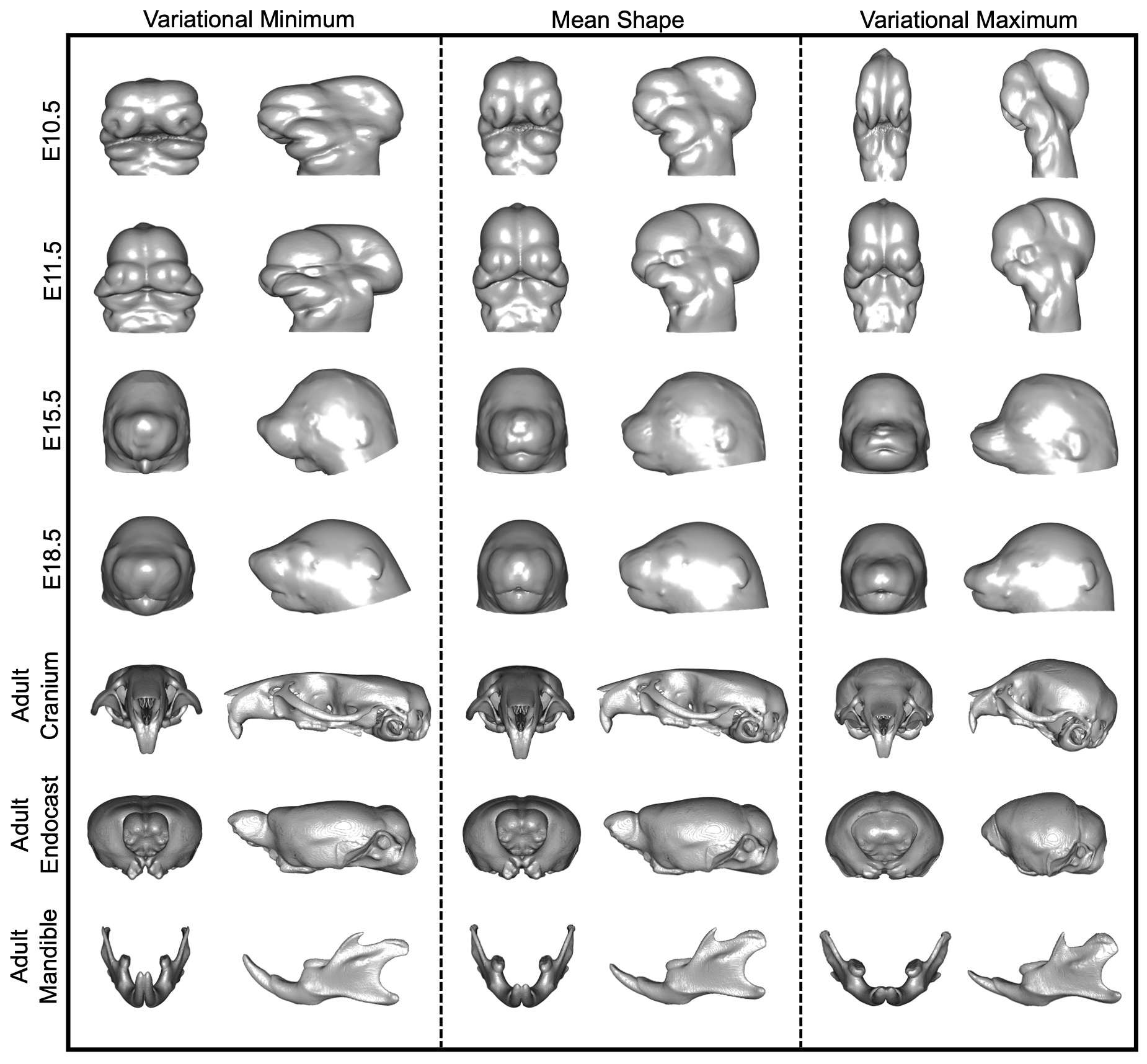
FaceBase is excited to announce the release of a large database of standardized mouse morphology data spanning numerous genotypes and developmental stages. This dataset, deposited by Jay Devine and Benedikt Hallgrimsson of the University of Calgary, encompasses one of the largest collections in FaceBase to date. And because the data are standardized, this allows for accurate comparisons with other datasets for analysis.
Here is the complete description from the MusMorph project page:
Complex morphological traits are the product of many genes with transient or lasting developmental effects that interact in an anatomical context. Mouse models are a key resource for disentangling such effects, because they offer myriad tools for manipulating the genome in a controlled environment. Unfortunately, phenotypic data are often obtained using laboratory-specific protocols, resulting in self-contained datasets that are difficult to relate to one another for larger scale analyses. To enable meta-analyses of morphological variation, particularly in the craniofacial complex and brain, we created MusMorph, a database of standardized mouse morphology data spanning numerous genotypes and developmental stages, including E10.5, E11.5, E14.5, E15.5, E18.5, and adulthood. To standardize data collection, we implemented an atlas-based phenotyping pipeline that combines techniques from image registration, deep learning, and morphometrics. Alongside stage-specific atlases, we provide rigidly aligned micro-computed tomography images, dense anatomical landmarks, and segmentations (if available) for each specimen (N=10,056). Our workflow is open-source to encourage transparency and reproducible data collection. The MusMorph data and metadata are stored across datasets in this project directory. The atlas files and project-wide metadata can be found in the project-wide dataset. The MusMorph scripts can be found on GitHub.
